On this page:
- Which JupyterLab environment should I choose?
- What packages are in the New Default Environment?
- How do I access RStudio?
- How can I install custom packages?
- How do I create a Conda environment?
- What is a Jupyter kernel?
- How do I restart my JupyterLab server?
- Can I recieve announcements for downtime and other critical information about LibreTexts/UC Davis JupyterHub?
- How do I distribute files to students?
- How do I download/upload files to my account?
For instructors and their students
Which JupyterLab environment should I choose?
After logging in to JupyterHub, you will be taken to a Server Options page where you may select your JupyterLab evironment. This will determine the languages, packages, and kernels available to you. You should continue with whichever option is selected by default unless you have a specific reason to do otherwise.
Each Jupyterhub environment is a different Docker image that contains all the packages, languages, and kernels you need. Docker images are basically snapshots of an entire computer system, so you are being served the exact same system state each time you boot a Jupyter environment, plus your personal files.
The New Default Environment is what most users should choose for general JupyterLab use. It is an updated version of what is now called Legacy Default Environment. In the new environment, all supported languages except for Octave and C++ have been upgraded. Depending on the language, some syntax might have changed, so be sure to test your code to see if it works on a newer version. Here's a list of all the languages we support and the version changes:
| Language | Old version | New version |
|---|---|---|
| Python | 3.6 | 3.7.8 |
| R | 3.6.1 | 4.0.2 |
| Julia | 1.1.0 | 1.5.1 |
| Octave | 4.2.2 | 4.2.2 |
| C++ (cling) | 0.6 | 0.6 |
| SageMath | 8.1 | 9.1 |
Starting with R 4.0, there has been a change to the behavior of data.frame() and read.table() calls. To maintain compatibility with previous code, you will want to add a stringsAsFactors=TRUE parameter to each of your data.frame() and read.table() calls.
Most of the packages included in this New Default Environment are at their latest version. Compared to Legacy Default Environment, a few packages have been removed in this new environment, which are listed below.
- pymol and ipymol: Requires extra conda channels and is not entirely open source software. Can be installed manually in a new conda environment.
- opty: Requires Python <=3.6. Can be installed manually in a new conda environment with Python 3.6.
- rpy2: Requires R <= 4.0. Can be installed manually in a new conda environemt with R 3.6.
If you require the above packages or older versions of existing ones, you should use Legacy Default Environment or create a conda environment with your custom package versions. Please email us at jupyterteam@ucdavis.edu if you have concerns or issues with the new JupyterLab environment.
What packages are in the New Default Environment?
The environment is generated from the default environment repository. apt.txt is a list of all apt packages and environment.yml is a list of all conda and pip packages.
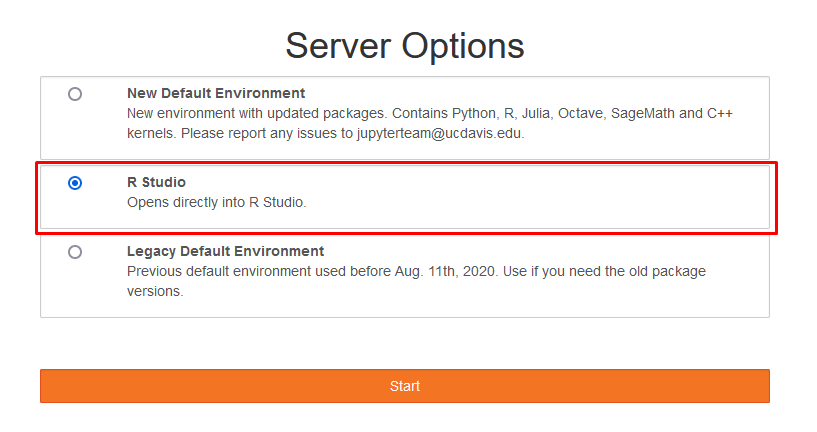
How do I access RStudio?
To open the RStudio interface, you can select the RStudio environment in the server options window when first starting your JupyterLab server. This will open a new JupyterLab tab which has an RStudio interface and access to all the same packages you would find elsewhere on your account.
How can I install custom packages?
You are free to install any additional packages available through conda's conda-forge channel by creating a conda environment, or by running conda install conda install directly in the (notebook) conda environment provided by default, but these packages will not be available when the server restarts.
If you cannot find your package or are uncertain how to install it, we suggest that you email us to provide them for you by default in the Jupyterhub environment. We can only support packages available from Ubuntu 18.04 APT repositories and from conda's conda-forge channel. Anyone can submit packages to both the APT and conda-forge repositories. Once packages are available there, email us with the package names and we will install them. Instructors wanting to create custom packages for their class will probably want to email us as well.
How do I create a Conda environment?
The best way to permanently install new packages onto your account is through conda environments. Conda environments are file directories which contain isolated collections of packages that you can install with the conda package manager (available by default in all our Jupyterhub environments). Using conda environments, you may also build Jupyter notebooks and consoles which contain only the programming language and packages that you need. Follow the steps below to create one.
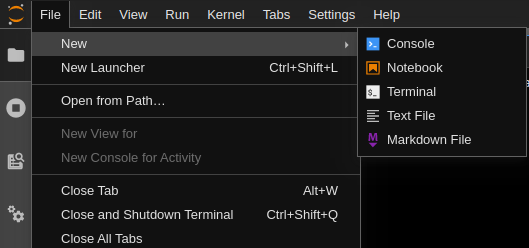
In the top left corner, open a terminal by going to File->New->Terminal or open a New Launcher (Ctrl+Shift+L) and select terminal from the 'other' section.
Within the terminal, use the command
conda create -n <your_env_name> <kernel> <packages>to create your environment. Below is an example that creates an environment calledscipkg, which contains theipykernelkernel along with the python packagesnumpy,pandasandmatplotlib. In general, you may include any packages which are available through conda.
Do not worry if you recieve a warning about newer versions of conda when running the
conda createcommand. Just proceed through the installation when prompted by pressingyor by initially runningconda createwith a-yflag at the end.After the conda environment has been built, activate it using
conda activate <your_env_name>. You should now see(your_env_name)at the beginning of the terminal prompt. This indicates that the conda environment is active. Note that the conda environment will only be active within that specific terminal session. To deactivate, runconda deactivate.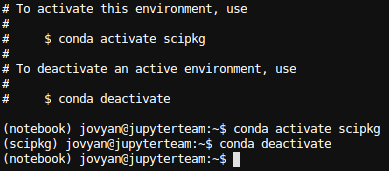
If you specified a Jupyter kernel then you may run your conda environment as a Jupyter notebook or console. Open the launcher (Ctrl+Shift+L) and select your conda environment from there. Using the
scipkgexample, the environment will appear asPython [conda env:.conda-scipkg]. You may have to wait a little bit for the icon to appear; refresh the webpage if needed.
Once the notebook opens, you can verify that you are using the proper conda environment by looking in the top right corner;

Now your conda environment is created! You may access it in the terminal by
conda activate <your_env_name>or as a notebook from the launcher.
Note that if you want to use your conda environment as a Jupyter notebook, you must install a Jupyter kernel along with your packages. Here's a list of common languages and their kernels which are supported through conda:
| Language | Jupyter kernel |
|---|---|
| Python | ipykernel |
| R | r-irkernel |
| C++ | xeus-cling |
| Octave | octave_kernel |
You may view all your created conda environments using conda env list. To remove an environment, use conda env remove -n <your_env_name>. For reference, all of your created conda environments are stored in /home/jovyan/.conda/envs/. You can learn more about managing and creating conda environments and more conda commands from the linked documentation.
Note that you have 10GB of storage available to your account. Some packages take up quite a bit of storage, so keep this in mind when you are creating conda environments.
What is a Jupyter kernel?
Jupyter kernels process the code you type and display the results to Jupyter notebook, console, or other tools. This typically means that each kernel supports a specific programming language and version, which is why you must include a kernel in your conda environment to run the environment as a notebook. As an example, if you have ever used JupyterLab locally for python programming then you have made use of the ipython kernel.
Kernels have a runtime which is independent of any given notebook, console, or terminal, and you can find more about managing kernels from the official JupyterLab documentation. There is also a Kernel tab at the top of your JupyterLab which will let you easily shutdown/restart kernels. You generally should not have to worry about this, but it may be useful for certain debugging purposes. A full list of Jupyter Kernels is available here. If you'd like us to support any additional kernels, go ahead and email us.
How do I restart my JupyterLab server?
On LibreTexts/UC Davis JupyterHub, you spawn servers that provide you with JupyterLab software and you execute your code using kernels. If you are actively running code and something unexpected occurs, restarting the kernel can often be an easy solution. However, sometimes the problem may be deeper than that, especially if some of your files were incorrectly modified. In this case, it could be a good idea to restart your server.
Go to File->Hub Control Panel and you will be brought to a page with "Stop Server" and "My Server" buttons. Press Stop Server and you should see that that the My Server button will dim; this means that the server is stopping. When the server has stopped (this should take less than 15 seconds), the webpage will prompt you with a "Start My Server" option. Press it, launch the server, and then you will be brought back to the Server Options page. Proceed with the JupyterLab environment of your choice, and you will now have a fresh server.
By default, your server will always shutdown after 1 hour of inactivity. All files which you wish to modify and save across restarts must be located in your /home/jovyan directory; files stored in other locations will not be saved. Think of these /home/jovyan user files as being stored in a cloud, and each time you start JupyterLab, we provide you with a brand new server which has downloaded your specific files. We backup these user files for disaster and recovery purposes, but you should preform your own backup service for version control as needed. For more technical information on how your files are stored, see the Jupyterhub for Kubernetes documentation here.
Can I recieve announcements for downtime and other critical information about LibreTexts/UC Davis JupyterHub?
If you wish to recieve updates from the JupyterTeam about possible downtime for LibreTexts/UC Davis JupyterHub, you can subscribe to our mailing list. Just enter your email and you will recieve emails whenever we notify Hub users about downtime and maintenance.
For instructors and their students
How do I distribute files to students?
The easiest way to distribute files to students for use on JupyterHub is through nbgitpuller. Nbgitpuller works by creating a link which automatically pulls files from a git repository and downloads them to a student's JupyterHub account. Since we already have nbgitpuller installed, all you need to do is create a github repository containing all the files you would like to share, and then fill in the Git Repository URL section of the nbgitpuller link generator with the URL of that online repository. Afterwards, simply share the generated link with your students and when they click on it, all of the files within the repository will be downloaded to their JupyterHub account. There are some nuances to the download process, so check this page if you are experiencing unexpected problems. If you are uncertain how to create a github repository, you can try this short tutorial. If you have never used git before and don't feel comfortable learning, Google Drive also offers you a great way to share your files.
How do I download/upload files to my account?
- The simplest way to upload files is through the built-in tool. With the File Browser tab selected, click on the Upload Files button. This will open a prompt to select the desired file to upload.
- If the file is not available locally, the
curlorwgetcommands can be used. To start, enter a terminal instance. To just download the file, the command usage for wget iswget [link to file]. For curl, it would becurl [link to file] -o [output file name]. Either of these will download the file into JupyterLab. It is encouraged to look into the greater functionalities of both tools to make the best use of them.